PhD Public Defense, Nasir Uddin
Title: Developing Composite Molecular Systems Network of Aggression
by Nasir Uddin
PhD Public Defense
Advisors:
Dr. Imran Rauf and Dr. Mushtaq Hussain (DOW)
External Examiners:
1. Dr. Ishrat Jabeen, School of Interdisciplinary Engineering & Sciences, NUST, Islamabad
2. Dr. Zaheer Ul-Haq Qasmi, Panjwani Center for Molecular Medicine and Drug Research, ICCBS, Karachi
Date: Friday, Feb 17, 2023 at 3:00pm
Venue: Conference Room, Tabba Academic Block, Main Campus, IBA Karachi
Abstract
Aggressive behavior is regulated by the interplay of several biomolecules that work in a variety of permutations of combination for the expression of a trait. This makes the identification of the key molecular components that underpin aggression difficult. Herein, we have used a systems biology approach to develop a composite system network in order to explore key proteins involved in the incidence and progression of aggression. In summary, an extensive literature survey yielded a total of 383 genes in this study. Stringent exclusion criteria were employed to weed out comorbidities like cancer and other pathological and physiological ailments, redundant and orphan genes. Resultantly, a single network of 351 genes was developed. The sparse protein-protein interaction network is composed of 383 nodes and 3,472 edges. The largest connected sub-network coalesced 351 genes of the sparse network, whereas 32 genes remained orphans. The subsequent network was searched for molecular complexes and ten modules were identified. Each module was subjected to enrichment and ontological analysis to identify important pathways associated with each module. Finally, novel candidate genes for aggression were identified by connecting different modules on an incremental basis.
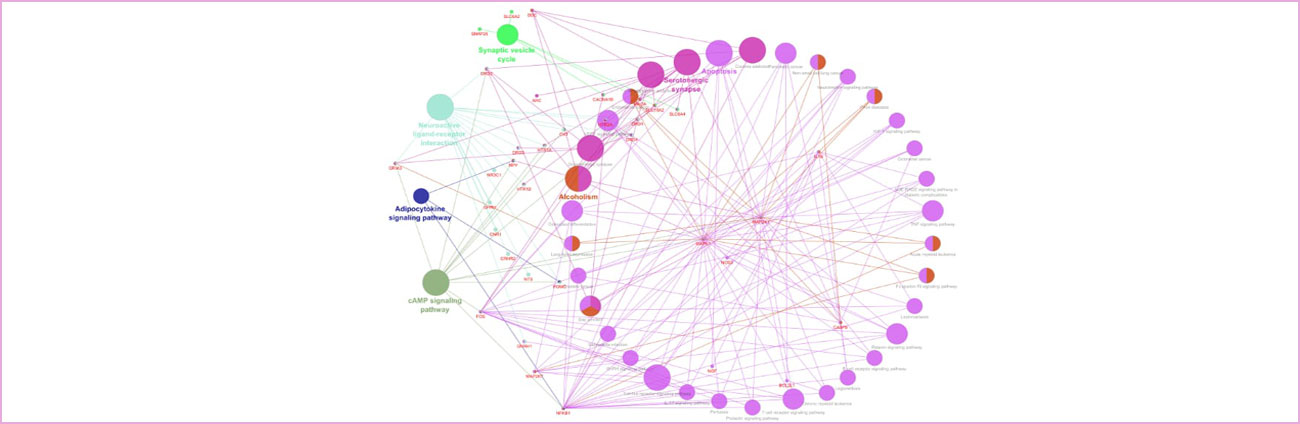
A dense network was created using the genes of the sparse network in addition to the nominally associated genes via genome wide association studies. This formed an interactome of 748 nodes, 6042 edges. The interactome had 49 unconnected components. The largest sub-network has 698 nodes and 6040 edges. The enrichment analysis of the sparse network showed that the biological processes these genes are mainly composed of are: behavior, synaptic signaling and trans-synaptic signaling. The molecular functions highly enriched were protein binding, neurotransmitter receptor activity, signaling receptor binding.
The sparse network modular analysis resulted in ten modules. The top three modules enrichment analysis revealed that the majority of the genes belonging to these modules belong to neuroactive ligand-receptor interaction, cAMP signaling pathway, serotonergic synapse, Amphetamine addiction, Neuroactive ligand-receptor interaction, Cocaine addiction, Neuroactive ligand-receptor interaction and Insulin secretion.
Individual modules were also analyzed to identify the genes that are involved in the crosstalk amongst different pathways within a module. In the highest scoring module DRD1, DRD2, NFKB1, MAPK1, and NPY play roles in the crosstalk of five different pathways, whereas HTR1A and HTR1B influence three pathways. TH was found to be influencing amphetamine addiction, tyrosine metabolism and folate biosynthesis in the second ranked module. The sparse network was analyzed for hub genes and BDNF, FOS, CREB1, IL6 and GRIN2B were found to be the most promiscuously interacting genes.
Potential novel genes were predicted based on additional genes invocation through the STRING Database. These genes were ranked based on their frequency of appearance in interaction with random modules. Furthermore, potential driver genes were also predicted using the dense network genes involved in module bridging using the shortest path approach. A statistically significant difference was observed in the length of shortest paths between dense network genes and nominally associated genes identified through GWAS studies.
Based on the expression data of normal subjects' correlation interaction networks for different age groups revealed that common subnetwork of the genes involved in the regulation of aggressive behavior. This analysis revealed the promiscuously interacting GRIN2B, and NPY are among the top interacting genes in the sparse and dense networks. Besides that, these genes were also among the top in module bridging and betweenness measures. Suggesting NPY and GRIN2B are potentially the most important drivers of aggressive behavior.
The data points to the engagement of several modules of the variable population in the development of aggression. Certain molecules appeared to be highly promiscuous in their interaction representing the potential driver and/or hub genes. In addition, the molecular interaction sites explored by molecular docking could be exploited to design a therapeutic intervention to ameliorate aggression.